- Home
- Labs
- Pierce Lab
- Projects
- Pericentral Retinitis Pigmentosa
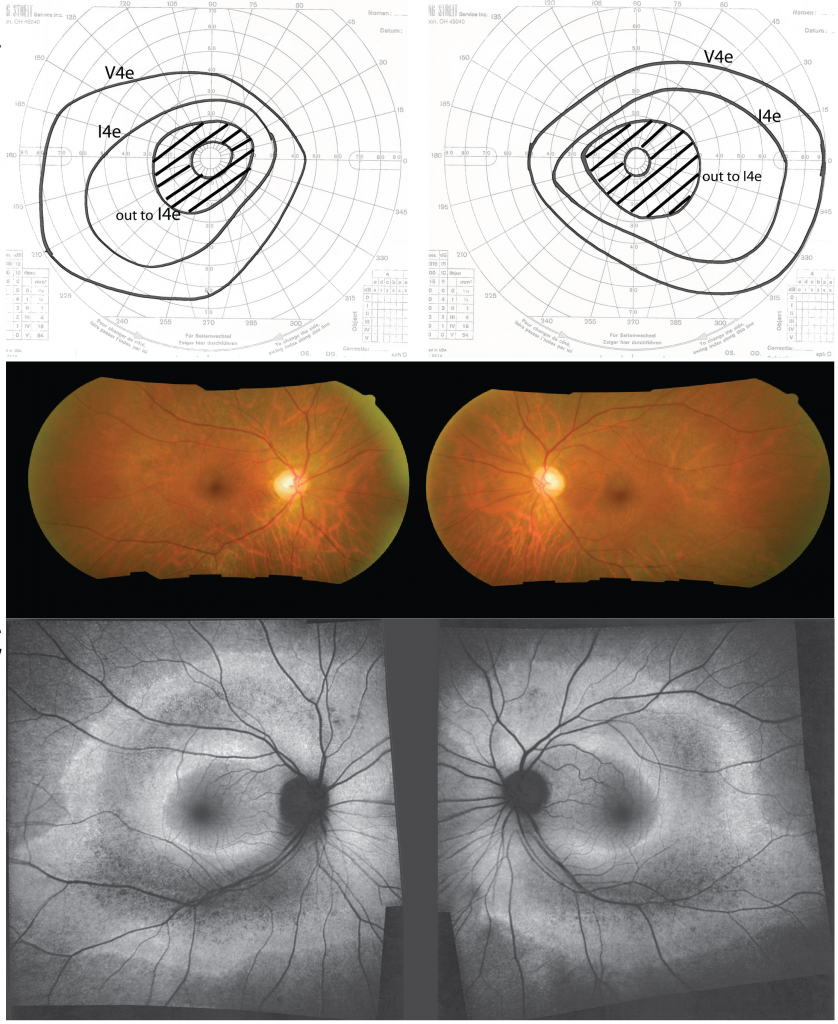
Visual fields (top), color fundus phtotos (middle), and autofluorescence imaging (bottom) of a patient with pericentral retinitis pigmentosa.
Pericentral retinitis pigmentosa (RP) is an atypical form of RP that affects the near-peripheral retina first and tends to spare the far periphery. This study was performed to further define the genetic basis of this phenotype. We identified a cohort of 43 probands with pericentral RP based on a comprehensive analysis of their retinal phenotype. Genetic analyses of DNA samples from these patients were performed using panel-based next-generation sequencing, copy number variations, and whole exome sequencing (WES). Mutations provisionally responsible for disease were found in 19 of the 43 families (44%) analyzed. These include mutations in RHO (5 patients), USH2A (4 patients), and PDE6B (2 patients). Of 28 putatively pathogenic alleles, 15 (54%) have been previously identified in patients with more common forms of typical RP, while the remaining 13 mutations (46%) were novel. Burden testing of WES data successfully identified HGSNAT as a cause of pericentral RP in at least 2 patients, suggesting it is also a relatively common cause of pericentral RP. While additional sequencing might uncover new genes specifically associated with pericentral RP, the current results suggest that genetically pericentral RP is not a separate clinical entity, but rather is part of the spectrum of mild RP phenotypes.
