- Home
- Labs
- Pierce Lab
- Projects
- NMNAT1 Leber Congenital Amaurosis (LCA)
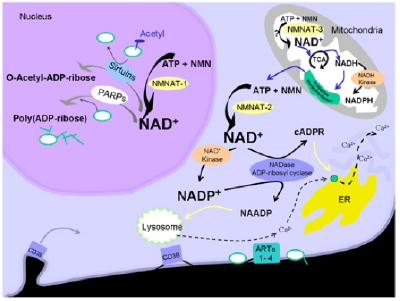
We and others recently reported that mutations in the NMNAT1 gene are a common cause of Leber congenital amaurosis (LCA), accounting for ~5% of cases (1-5). NMNAT1 encodes an enzyme that is essential for generating NAD+ (6). Three mammalian NMNAT isoforms that are encoded by different genes have been identified within distinct cellular compartments, where NMNAT1, 2 and 3 localize, respectively, to the nucleus, Golgi complex, and mitochondria (7,8). NMNAT1 regenerates nuclear NAD+ to support DNA metabolism and cell signaling, whereas NMNAT3 regenerates mitochondrial NAD+ for use in cellular energetics (Figure 1) (8).
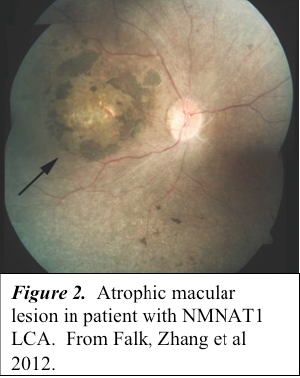
The identification of NMNAT1 as an LCA disease gene raises the question of how mutations in a widely expressed NAD+ biosynthetic protein lead to a retina-specific phenotype. The answer to this question is of particular interest because the majority of patients with NMNAT1-LCA have atrophic macular lesions, suggesting that macular cones are especially dependent upon NMNAT1 activity (Figure 2) (2-5). Of interest, NMNAT1 is the principle component of the Wallerian degeneration slow (WldS) fusion protein which has been found in mice to permit injured axons to survive ten times longer than normal (9-11).
We are currently investigating the pathogenesis of NMNAT1 disease via biochemical studies and developing an adeno-associated virus (AAV)-mediated gene augmentation therapy as a potential treatment. Experiments are being performed in Nmnat1 mutant mice (12) that harbor a human mutation and recapitulate key features of NMNAT1-LCA. Please click here for our Nature Genetics publication on NMNAT1 and click here for our American Journal of Pathology publication describing the NMNAT1 mouse model.
References
1. den Hollander AI, Black A, Bennett J, Cremers FP.Lighting a candle in the dark: advances in genetics and gene therapy of recessive retinal dystrophies. The Journal of clinical investigation. 2010;120(9):3042-53. PMCID: 2929718.
2. Falk MJ, Zhang Q, Nakamaru-Ogiso E, Kannabiran C, Fonseca-Kelly Z, Chakarova C, Audo I, Mackay DS, Zeitz C, Borman AD, Staniszewska M, Shukla R, Palavalli L, Mohand-Said S, Waseem NH, Jalali S, Perin JC, Place E, Ostrovsky J, Xiao R, Bhattacharya SS, Consugar M, Webster AR, Sahel JA, Moore AT, Berson EL, Liu Q, Gai X, Pierce EA.NMNAT1 mutations cause Leber congenital amaurosis. Nature Genetics. 2012;44(9):1040-5.
3. Koenekoop RK, Wang H, Majewski J, Wang X, Lopez I, Ren H, Chen Y, Li Y, Fishman GA, Genead M, Schwartzentruber J, Solanki N, Traboulsi EI, Cheng J, Logan CV, McKibbin M, Hayward BE, Parry DA, Johnson CA, Nageeb M, Poulter JA, Mohamed MD, Jafri H, Rashid Y, Taylor GR, Keser V, Mardon G, Xu H, Inglehearn CF, Fu Q, Toomes C, Chen R.Mutations in NMNAT1 cause Leber congenital amaurosis and identify a new disease pathway for retinal degeneration. Nature Genetics. 2012;44(9):1035-9.
4. Chiang PW, Wang J, Chen Y, Fu Q, Zhong J, Yi X, Wu R, Gan H, Shi Y, Barnett C, Wheaton D, Day M, Sutherland J, Heon E, Weleber RG, Gabriel LA, Cong P, Chuang K, Ye S, Sallum JM, Qi M.Exome sequencing identifies NMNAT1 mutations as a cause of Leber congenital amaurosis. Nature Genetics. 2012;44(9):972-4.
5. Perrault I, Hanein S, Zanlonghi X, Serre V, Nicouleau M, Defoort-Delhemmes S, Delphin N, Fares-Taie L, Gerber S, Xerri O, Edelson C, Goldenberg A, Duncombe A, Le Meur G, Hamel C, Silva E, Nitschke P, Calvas P, Munnich A, Roche O, Dollfus H, Kaplan J, Rozet JM.Mutations in NMNAT1 cause Leber congenital amaurosis with early-onset severe macular and optic atrophy. Nature Genetics. 2012;44(9):975-7.
6. Belenky P, Bogan KL, Brenner C.NAD+ metabolism in health and disease. Trends in Biochemical Sciences. 2007;32(1):12-9.
7. Berger F, Lau C, Dahlmann M, Ziegler M.Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J Biol Chem. 2005;280(43):36334-41.
8. Lau C, Niere M, Ziegler M.The NMN/NaMN adenylyltransferase (NMNAT) protein family. Frontiers in bioscience : a journal and virtual library. 2009;14:410-31.
9. Coleman MP, Freeman MR.Wallerian degeneration, wld(s), and nmnat. Annual review of neuroscience. 2010;33:245-67.
10. Lunn ER, Perry VH, Brown MC, Rosen H, Gordon S.Absence of Wallerian Degeneration does not Hinder Regeneration in Peripheral Nerve. The European journal of neuroscience. 1989;1(1):27-33.
11. Conforti L, Tarlton A, Mack TG, Mi W, Buckmaster EA, Wagner D, Perry VH, Coleman MP.A Ufd2/D4Cole1e chimeric protein and overexpression of Rbp7 in the slow Wallerian degeneration (WldS) mouse. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(21):11377-82. PMCID: 17208.
12. Greenwald, S.H., Charette, J.R., Staniszewska, M., Krebs, M.P., Bowl, M.R., Shi, L.Y., Brown, S.D.M., Stone, L., Liu, Q., Hicks, W.L., Nishina, P.M. & Pierce, E.A. Mouse models of NMNAT1-Leber congenital amaurosis (LCA9) recapitulate key features of the human disease. American Journal of Pathology. 2016, 186(7): 1925-1938.