- Home
- Publication
- RETINAL Transcriptome
RETINAL Transcriptome
 June 3, 2017
June 3, 2017
Original work (2013)
View Data in Genome Browser
PROJECT DETAILS
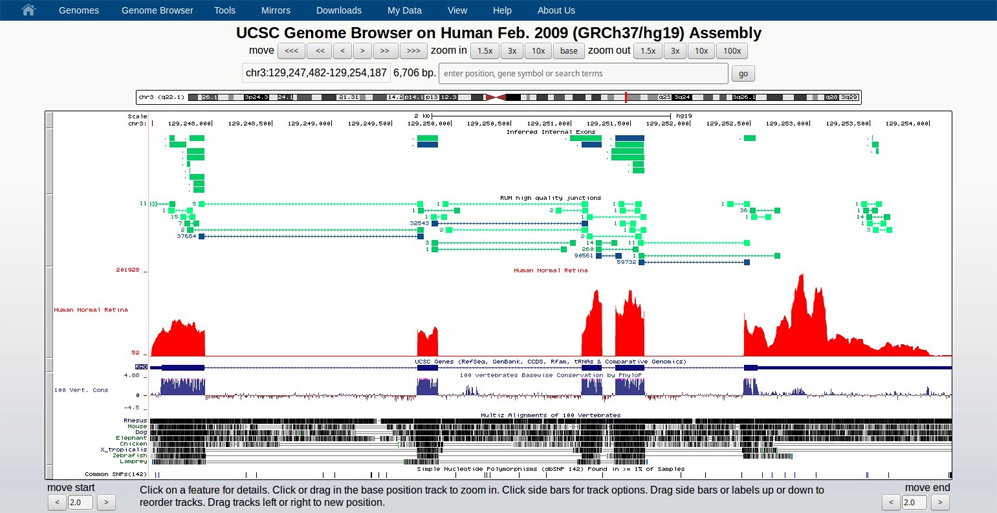
The human retinal transcriptome data, available on the UCSC Genome Browser, was derived by performing RNA-Seq on 3 normal retina samples. Over 300 million 101 bp paired-end reads were aligned using the RNA-Seq Unified Mapper . These data are available as a resource to the research community and can be used to identify novel alternative splicing events, relative gene expression levels, as well as novel gene expression.
Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genomics 2013, 14:486
UCSC genome browser link:
Human Normal Retina Transcriptome (2013)
Updated beta version (June 2019)
View Data in Genome Browser
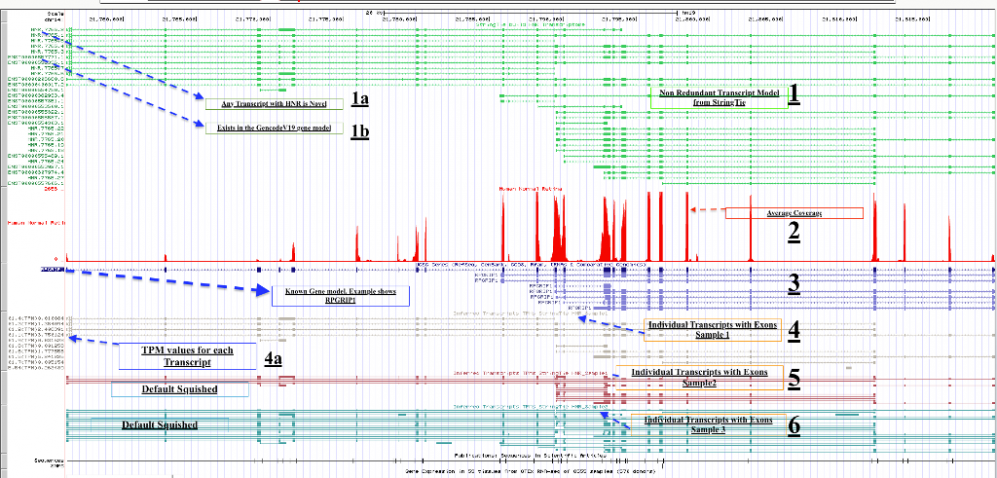
1) Shows the non redundant gene mode. This made after combining the samples. When we do that, we loose coverage. Hence that information is shown (please keep reading) in a different way (at this time, later will find new ways).
1a) Anything marked with HNR* is novel
1b) Anything with ENST* is annotation in the known gene model
2) Average coverage from the three samples
3) Known gene model (RefSeq)
4) Transcripts from human normal retina Sample1. Please take note towards the left, you will see that name contains TPM value. Its done in such a way that TPM values are visible. The name here will NOT match names at the top. The names are not retained when the data is merged
What you have TPMs for each transcript in the given sample. Now you can judge which transcript will be used to make a final transcripts. Higher the TPM the better?
5) Transcripts from human normal retina Sample2. By default this is in squished format. Please scroll below and make it “full”. TPMs are shown in the name of the left.
6) Transcripts from human normal retina Sample3. By default this is in squished format. Please scroll below and make it “full”. TPMs are shown in the name of the left.
Updated beta version (June 2019):
Human Normal Retina Transcriptome (BETA 2019)
IGV session (June 2019):