New Findings Published in BMC Genomics
May 1, 2017OGI Researcher Michael Farkas, Eric Pierce and colleagues publish groundbreaking findings in BMC Genomics; most thorough description of gene expression in the human retina to date.
BOSTON (July 18, 2013) – Investigators at Massachusetts Eye and Ear and Harvard Medical School have published the most thorough description of gene expression in the human retina reported to date. In a study published today in the journal BMC Genomics, Drs. Michael Farkas, Eric Pierce and colleagues in the Ocular Genomics Institute (OGI) at Mass. Eye and Ear reported a complete catalog of the genes expressed in the retina.
The retina is the neural tissue in the back of the eye that initiates vision. It is responsible to receiving light signals, converting them into neurologic signals and sending those signals to the brain so that we can see. If one thinks of the eye as a camera, the retina in the “film” in the camera. For these studies, the investigators used a technique called RNA sequencing (RNA-seq) to identify all of the messenger RNAs (mRNAs) produced in the human retina. The resulting catalog of expressed genes, or transcriptome, demonstrates that the majority of the 20,000+ genes in the human body are expressed in the retina. This in itself is not surprising, because the retina is a complex tissue comprised of 60 cell types.
In a more surprising result, Dr. Farkas and colleagues identified almost 30,000 novel exons and over 100 potential novel genes that had not been identified previously. Exons are the portions of the genome that are used to encode proteins or other genetic elements. The investigators validated almost 15,000 of these novel transcript features and found that more than 99 percent of them could be reproducibly detected. Several thousand of the novel exons appear to be used specifically in the retina. In total, the newly detected mRNA sequence increased the number of exons identified in the human genome by 3 percent.
“While this may not sound like a lot, it shows that there is more to discover about the human genome, and that each tissue may use distinct parts of the genome,” said Dr. Pierce, Director of the OGI and the Solman and Libe Friedman Associate Professor of Ophthalmology, Harvard Medical School.
This work is valuable to help scientists understand how the retina worksand how it is affected by disease. For example, Dr. Pierce and colleagues in the OGI study inherited retinal degenerations, which are common causes of vision loss. These diseases are caused by misspellings or mutations in genes that are needed for vision. To date, investigators have identified more than 200 retinal degeneration disease genes, but still can’t find the cause of disease for up to half of the patients affected by these disorders. Identification of new exons used in the retina may help find the cause of disease in these patients. The transcriptome data can be viewed via the OGI website at http://oculargenomics.meei.harvard.edu/index.php/ret-trans.
Identifying the genetic cause of patients’ retinal degeneration has become especially important with the recent success of clinical trials of gene therapy for RPE65 Leber congenital amaurosis (LCA). As a follow-up to these initial proof-of-concept trials, clinical trials of gene therapy for four other genetic forms of inherited retinal degeneration are currently in progress. Further, studies in animal models have reported successful gene therapy for multiple additional genetic types of IRD. There is thus an unprecedented opportunity to translate research progress into provide sight preserving and/or restoring treatment to patients with retinal degenerative disorders.
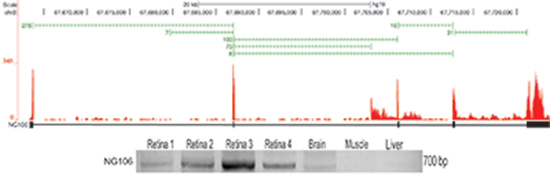
Identification and tissue distribution of novel genes expressed in the retina. RNA-Seq was performed on three normal human retina samples. Over 300 million reads were aligned to the human genome using the RNA-Seq Unified Mapper (RUM). Using the novel splice junctions (green bars) and coverage data (red), transcripts with 3 or more novel exons located in intergenic regions were identified as putative novel genes (transcript models in black). An example of a novel six exon gene (NG106) that contains an open reading frame and is believed to be protein coding. A majority of the novel genes, however, are not believed to be protein coding, suggesting they are a class of regulatory non-coding RNAs known as long intergenic non-coding RNAs (lincRNAs). Primers were designed in the putative terminal exon regions of the novel genes, and subject to RT-PCR and Sanger sequencing for validation of expression. NG106 was not found in the liver and muscle, suggesting its expression may be limited to specific tissues such as the retina.
The transcriptome data can be viewed on the UCSC Genome Browser at http://oculargenomics.meei.harvard.edu/index.php/ret-trans/110-human-retinal-transcriptome.
